
The current trajectory for crop yields is insufficient to nourish the world’s population by 2050 1 . Greater and more consistent crop production must be achieved against a backdrop of climatic stress that limits yields, owing to shifts in pests and pathogens, precipitation, heat-waves and other weather extremes. Here we consider the potential of plant sciences to address post-Green Revolution challenges in agriculture and explore emerging strategies for enhancing sustainable crop production and resilience in a changing climate. Accelerated crop improvement must leverage naturally evolved traits and transformative engineering driven by mechanistic understanding, to yield the resilient production systems that are needed to ensure future harvests.

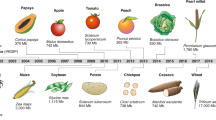
The Green Revolution of the 1960s enabled a steep increase in the yields of major staple grain crops (wheat, corn and rice) to address the caloric needs of an increasing global population. This was accomplished through elite variety breeding, hybrid crop development, fertilizer application and advances in management through substantial public investment 2 . The consequent rise in food security benefitted many regions of the world and improved agricultural development (particularly in India and southeast Asia), reducing poverty and malnourishment.
By the 1980s, molecular and transformation technologies propelled the delivery of the first bioengineered genes into plant genomes. Currently, the most widely adopted genetically modified traits are resistance to herbicides and insects in crops with large markets (maize, soybean, cotton and Brassica napus (canola)). Although herbicide- and insect-resistance traits greatly lessened soil tillage and insecticide use, respectively, they require careful management to avoid natural selection of resistance in weeds or pests 3,4 . Despite engineered traits with clear benefits to farmers and end-users (including virus-resistant papaya 5 , drought-tolerant corn 6 , rice 7 and bananas 8 fortified with provitamin A, non-browning apples 9 and low-acrylamide potatoes 10 ), the acceptance of genetically modified traits is equivocal in some countries, and the cultivation of genetically modified crops is largely banned in the European Union.
Future food security will require reducing crop losses due to environmental factors, including climate change, as well as transformative advances that provide major gains in yields. More recent genomic technologies have expedited breeding and trait development for increased environmental resilience and productivity. Genetic diversity is now readily explored at nucleotide-scale precision, using genome-wide association studies and other gene-mapping methods paired with advanced phenotyping systems. The identification of loci that contribute to traits, coupled with molecular-marker-assisted breeding, enables the rapid selection of new genetic combinations in elite varieties. Complementary to breeding approaches, advances in the spatial and temporal regulation of engineered genes and pathways are increasingly accelerated by the targeted editing of genomes using CRISPR–Cas technology. A greater understanding of plant mechanisms that increase yields in variable environments is essential to drive the necessary gains in crop improvement, which can be fuelled by genetic diversity and implemented by genome-scale breeding, finely-tuned gene engineering and more-precise agronomic management practices.
Despite the marked effect of the Green Revolution on food security, there were uneven consequences for human nutrition, the resilience of crops to stress, and the environment 2 . Asian populations benefitted from the increased production of staple grains, and the adoption of irrigation across vast areas 2,11 . The limited rise in food security in sub-Saharan Africa and other impoverished areas can be traced to geographically skewed support and a lack of investment in orphan crops 2 . An unintended consequence has been that fruits and vegetables rich in macronutrients have been displaced by calorie-rich and higher-value grain crops in some areas 2 . Moving forward, an increased production of nutrient-rich vegetable, pulse, tuber and cereal crops, and a broadening of the global reach of agricultural advances, is necessary to achieve food and nutritional security 12 .
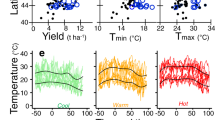
The increasing frequency of debilitating heat-waves, droughts, torrential rains and other weather extremes experienced across the globe negatively affects agricultural productivity, and is projected to do so 1,13 (Fig. 1). Climatic constraints can occur independently or together (as with heat and aridity), and in either case reduce the level of productivity that is predicted for a well-managed environment (the yield potential). An unanticipated consequence of the development of high-yield varieties for locations with advanced cultivation practices has been a loss of genetic variation that is associated with resilience to suboptimal environments. It is imperative to breed crops that carry a diversity of resistance genes and/or to plant a diversity of varieties, as this approach minimizes the ability of pathogens to overcome resistance 14 . An increasing occurrence of extreme weather events, together with dire projections of climate change, makes the improvement of crop resilience to environmental (abiotic) and pathogen (biotic) stress of paramount importance for feeding a growing global population.
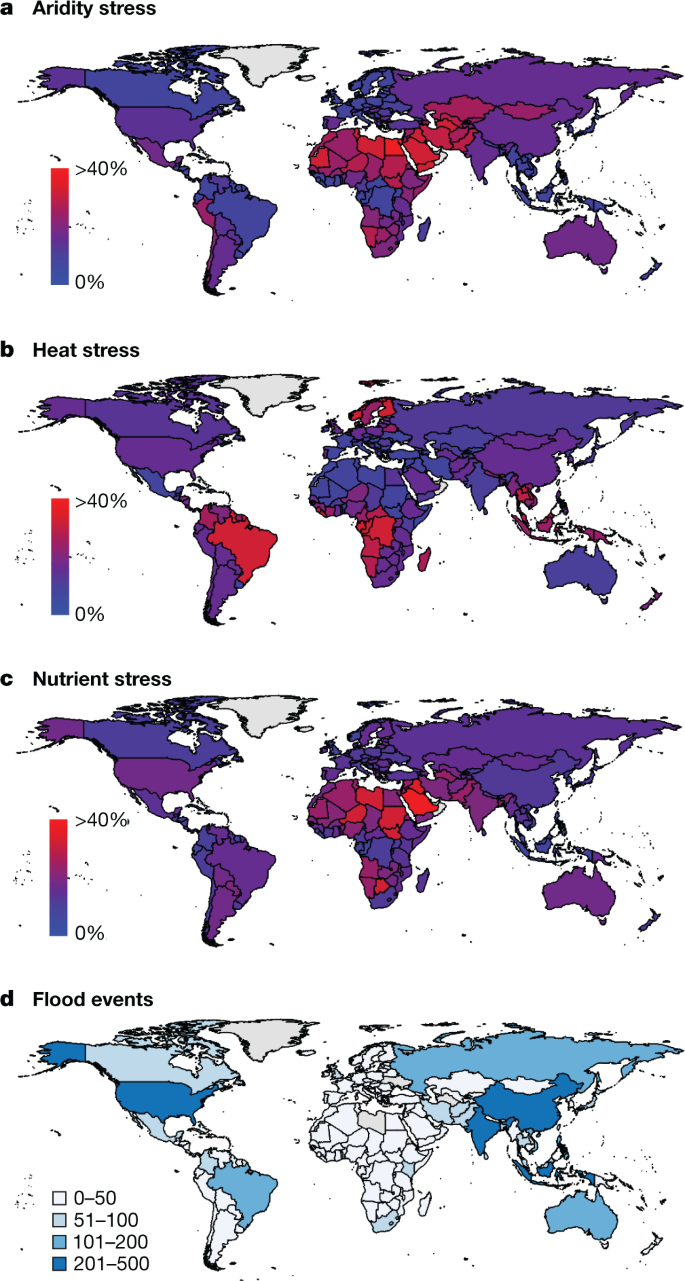
The combination of high-yield crop varieties and the widespread use of inorganic fertilizers markedly improved crop production, with clear benefits in terms of food security 15 . This has translated to excessive anthropogenic release of reactive nitrogen 16 and phosphate 17 into the environment. Inorganic fertilizers have pushed the global nitrogen and phosphorus cycles well beyond their estimated safe operating space 18 , with considerable negative effects on biodiversity, human health and the atmosphere 19 . Their use presents a paradox, as the optimization of plant nutrition stabilizes yields and has helped to reduce expansions in crop area in light of population growth, yet nitrogenous fertilizers contribute substantially to the greenhouse gas emissions that promote climate change 19 .
The agriculture of the next decades must satisfy demands for nutritious food, fibre and animal feed in a highly variable climate, and also mitigate the effects of agriculture on the environment. This is a tall order. Key to addressing the challenge is a deeper understanding of genetic variation and the molecular, cellular and developmental pathways by which plants dynamically respond to and interact with their environment and pathogens, while maintaining growth, efficiency of nutrient use and fitness. New crop varieties ideally will have genetic combinations that alleviate losses from the multiple environmental and pest constraints that are encountered during the crop lifecycle in a farmer’s field. An important emerging and non-trivial goal is to optimize the efficiency of photosynthesis, water and nutrient use, including the fostering of beneficial interactions between plants and microorganisms that can promote nutrient acquisition. The integration of mechanistic understanding, genetic variation and genome-scale breeding towards technological solutions will be essential. Here we review advances and emerging directions within the plant sciences that may bolster yield-defining traits and resilience (Fig. 2, Box 1).
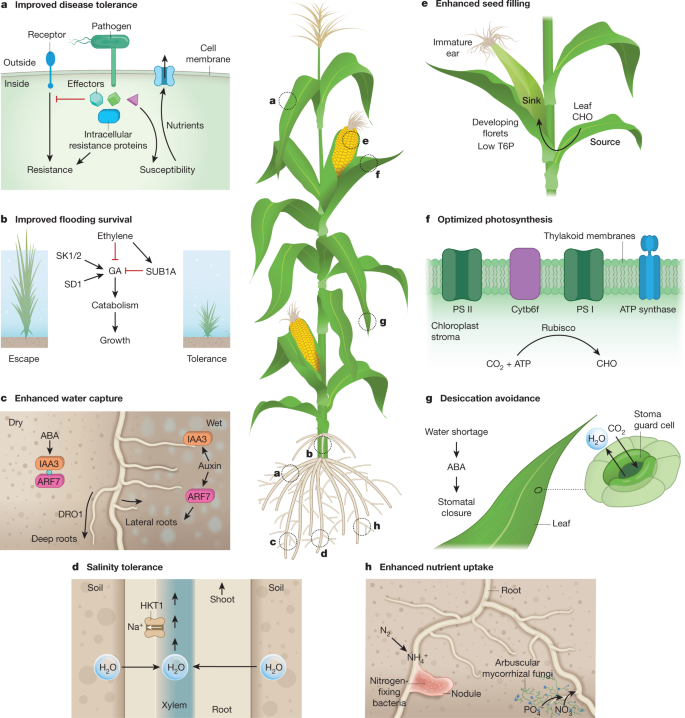
This Review discusses several traits that are essential for crop performance, including the genetic variation and plasticity that are relevant for improvement (left) and advanced and emerging approaches for addressing trait improvements (right). Stress resilience coupled with high yields is aided by hardwired traits and temporal responses to a dynamic environment. Opportunities for improvement include capturing natural genetic variation, the functional characterization of genes, the manipulation of endogenous or transferred genes with appropriate regulatory control, the development of low-cost and safe small molecules that can be delivered to plants before stress or during recovery, and improved plant health through interactions with symbiotic microorganisms.
Yield-defining traits
Shoot traits and plasticity
Inflorescence architecture and fertility
Stomatal movement and density regulation
Assimilate loading and partitioning
Root traits and plasticity
Architecture and anatomy
Nutrient acquisition and use efficiency
Stress resilience
Drought, salinity, flooding and extreme temperatures (abiotic)
Pests and pathogens (biotic)
Tempered response to minimize growth penalty
Opportunities
Natural genetic variation
Stress resilience and recovery mechanisms
Gene engineering and editing
Spatial, temporal and inducible control of genes and networks
Improving protein function, targeting and turnover
Enhancing metabolite pathway and flux
Introducing synthetic traits
Beneficial soil and leaf microbiome
Seeding and supplementation
Attraction of beneficial microorganisms
Small-molecule delivery
Sensor use for crop management
Cellular, organ, canopy and remote
The reliance on major pathogen-resistance genes bred into crop monocultures provides short-term protection against diseases, as seen in the boom-and-bust cycles of resistance over the past century and in the spread of new diseases across continents. A more complete molecular-genetic framework now exists for general and specific resistance to microbial pathogens mediated by a two-layered receptor signalling system (Fig. 2a). At the plant cell surface, families of pattern-recognition receptors register the presence of microorganisms 20 . Inside host cells, large panels of nucleotide-binding domain leucine-rich-repeat receptors (NLRs) detect activities of invasive pathogenic strains 21 . Advances in elucidating receptor–pathogen recognition and activation mechanisms at a protein-structural level provide strategies towards the rational design of receptor proteins that are tailored to intercept broader or alternative disease agents 21,22 . Newly engineered resistance traits can then be transferred to elite varieties of crops to confer resistance against modern diseases. There have been notable successes in the inter-family transgenic transfer of a pattern-recognition-receptor gene to potato, tomato, Medicago, wheat and rice 20,23 , indicating that surface receptors that are restricted to particular plant lineages can confer immunity in unrelated species. Transfer of the wheat Pm3e resistance gene against powdery mildew to a susceptible wheat variety has produced effective mildew resistance in field trials 24 . The engineering of pathogen-induced translational control of a key Arabidopsis immunity component in rice 25 has provided promising disease-resistance benefits in initial crop field trials, apparently without a yield penalty. The incorporation of new surface- and intracellular-receptor recognition and signal transduction modules into crops is also on the horizon, building on knowledge of receptor functional partnerships and resistance network architectures 21,26 . Success in this area—especially as climates change—will require tight immune-receptor control, which can require co-evolved factors 27,28 , because modified receptors without requisite constraints are often mis-activated and cause necrosis and reduced plant health. Knowledge in this arena may also lead to strategies that lessen food spoilage by pathogens after harvesting.
With increased access to diverse genetic variation found in crops and natural populations of wild relatives 14 , the door is open to recovering disease-resistance traits; many of these are encoded by genes for pattern recognition and NLRs that were lost during domestication, or that have evolved independently in different plant lineages 14,29 . Advances in genome sequencing and assembly technologies, coupled with new methods for capturing near-complete immune-receptor gene panels from complex genomes, hold promise for attaining sustainable disease resistance 30,31,32 . Merging these approaches with genome-wide association studies taps into immune-receptor gene variants that have adapted to local environments and pathogen populations to help to increase the resilience of future crops. The stacking (or ‘pyramiding’) of several resistance genes with different recognition spectra and environmental optima into a single background is now a credible strategy for achieving more durable disease resistance. Nevertheless, assembling appropriate gene combinations in elite varieties of crops remains a challenge.
Investigations of plant genomes within and across species provide insight into the evolutionary forces that have shaped the architecture and function of genes related to immunity. This will aid the design of new resistance traits 33 . The isolation and characterization of genes associated with disease susceptibility in the host has also gained prominence 34 . The proactive deployment of modified susceptibility genes in crops will become possible as geographical sampling of pathogen genomes and populations increases 35,36 . A recessive barley mildew resistance locus o (mlo) mutant that breeders have successfully used for 75 years against powdery mildew disease has been engineered into hexaploid wheat using mutagenesis by transcription-activator-like effector nuclease, or by combining mutations selected in three wheat MLO loci 37,38 . Thus, the characterization and manipulation of host–pathogen infection processes for the establishment of disease can generate novel resistance mechanisms in crops that are not necessarily found in natural populations. As a cautionary note, some newly engineered crop lines have displayed unexpected phenotypes and vulnerabilities to disease 39 , which underscores the need for rigorous performance testing of new material in field settings over multiple seasons.
Given the current trends in climate, attaining durable resistance in high-yield crops will require a greater knowledge of pathogen population dynamics and plant host responses to temperature. The alarming spread of devastating disease agents—such as the bacterium Xylella fastidiosa that attacks olives and woody crops in southern Europe, or the ug99 strain of stem rust fungus Puccinia graminis f. sp. tritici that affects wheat across parts of Africa and Asia—is attributed in part to warmer climates, and presents a complex biogeographical and epidemiological problem 40,41 . Moreover, surface- and intracellular-immunity systems appear to respond differently to changes in temperature, owing to effects on the microorganisms and hosts that are not completely understood 42,43 . Although immunity mediated by intracellular receptors tends to be less efficient as temperatures increase, some resistance genes confer protection at higher temperatures 44,45 . Although these findings are reassuring, they highlight the need for more genotype × environment studies in crops to stabilize resistance in increasingly precarious climates.
Abiotic stresses associated with climate change that destabilize yields include flooding, drought, soil salinity and extreme temperatures (Fig. 1). Resilience mechanisms have been mobilized for crop improvement through the identification of genes that are associated with key traits and signal transduction pathways, followed by breeding or engineering 46,47 (Fig. 2b–f). Attaining resilience without affecting overall yield is a considerable challenge.
Floods regularly limit yields 48 . Rice is exceptionally resilient against flooding, yet over 30% of the acreage cultivated with rice suffers yield loss owing to plant submergence 49 . SUBMERGENCE 1 (SUB1), identified in a flooding-tolerant landrace of rice, encodes a cluster of genes for ethylene-responsive transcription factors, including the submergence-activated SUB1A-1 50 . The SUB1A transcription factor curbs the activation of genes that promote the breakdown of leaf sugars and starch (photo-assimilate) that would otherwise fuel growth to escape an inundation 51 (Fig. 2b). The introduction of SUB1A-1, through marker-assisted breeding, into high-yield varieties now provides an additional week or more of submergence tolerance—without compromising yields under non-submergence conditions 52 .
The submergence tolerance by growth quiescence provided by SUB1A-1 contrasts with the accelerated underwater elongation of the shoots of varieties of crops that are adapted to progressive seasonal floods in delta regions. Deepwater varieties invest photo-assimilate into the extension of submerged stem internodes (Fig. 2b). This requires the SNORKEL 1 (SK1) and SNORKEL 2 (SK2) genes that encode transcription factors that are similar to SUB1A 53 , as well as biosynthesis of the hormone gibberellin. Gibberellin biosynthesis involves a functional allele of SEMIDWARF1 (SD1) 54 , the gene that—when mutated—determines the short stature of Green Revolution rice 55 . This knowledge can improve yields in low-lying areas that affected by climate change.
The alleles of SUB1A, SK1, SK2 and SD1 that are key to flooding survival are found in wild Oryza species 56 , which indicates that they arose in ancestral populations in flooded ecosystems. Evolution has modified the same growth-response network involving the hormones ethylene and gibberellin to achieve submergence tolerance or escape in diverse species of wetland plants 48 . Pathways to improved flooding tolerance include manipulation of root traits associated with waterlogging tolerance that involve a conserved mechanism 48 and the oxygen-dependent turnover of SUB1A-1-like transcription factors, accomplished in several species 57,58,59,60 . There are other opportunities to protect yields in wet climates. Torrential rain and hail can cause yield losses of 50% or more, owing to premature pod shattering in oil crops. The identification of genes that control pod shattering in Arabidopsis 61 enabled the gene-targeted molecular breeding of optimized pod-shattering properties in canola that is now increasingly planted by farmers.
Drought and other dehydration or osmotic stresses (salinity and cold) stimulate the production of the hormone abscisic acid (ABA) in plants. Although the mechanisms of the initial sensing of osmotic stress and signalling in response to osmotic stress remain poorly understood, the elucidation of the ABA receptor and signal transduction mechanisms 62,63 has exposed new avenues for the enhancement of dehydration tolerance. This includes ABA receptor overexpression 64,65 and engineering to respond to exogenously sprayed small molecules 66,67 , the overexpression of signal transduction components 68,69,70 or the drought-driven repression of negative regulators of ABA signal transduction 69,71 .
ABA closes the adjustable stomatal pores on the leaf surface that allow gas exchange and thus reduces the water lost from plants during drought, but this response can be weak in crop varieties 69,72 . ABA also helps to regulate root growth in response to water availability, including inhibition of lateral root growth and enhancement of primary and secondary root growth. This developmental reprogramming allows roots to seek water. The DEEPER ROOTING (DRO1) gene of rice provides a deep root architecture in paddy fields, which bolsters yields under water-limited conditions 73,74,75 (Fig. 2c). The identification of the major loci that control root traits associated with drought resilience has proven challenging owing to their quantitative nature and low heritability, which requires sophisticated belowground phenotyping and analytical methods 76 . Yet roots grow laterally towards moisture in soil 77 . New roots that access moisture emerge only on the side of a root that is moist, as a modification of a key auxin-response transcription factor on the dry side of a root impedes the developmental program 78 (Fig. 2c). Knowledge such as this can inform strategies for the advanced breeding and engineering of improved resilience to drought, which continues to limit yields 79,80 .
Irrigation substantially expands growing seasons and increases crop yields in many regions. Salt (sodium chloride) gradually accumulates in irrigated soils and is toxic to most crops; sodium accumulation is particularly detrimental in leaves. Approximately 40% of irrigated lands worldwide are affected by increased salt levels, and expansion of soil salinization is a major threat to crop performance 47 .
Plants encode a sodium-transporter gene sub-family named HKT1 81 (high-affinity K + transporter) that provides protection from the over-accumulation of sodium in leaves 82 (Fig. 2d). HKT1 mediates the removal of sodium, mainly in roots, from the xylem 83,84 , the vascular conduits that transport water and nutrients from roots to leaves. Major quantitative trait loci that enhance salt tolerance in wheat, wheat relatives and rice possess distinct HKT1 alleles 47,83,85 . This knowledge has enabled the marker-assisted breeding of wheat with a higher salt tolerance, resulting in a yield improvement of 25% under salinity stress in field trials 85 . Beneficial alleles of HKT1 may enhance salinity tolerance in other species, as has been shown in rice 83 . It will be necessary to combine HKTs with other strategies to further boost salinity resistance as land salinization continues to rise. The effects of salinity on root development also need to be factored into intervention strategies 86,87 . Natural variation in transporter genes and their regulation has also provided field-tested solutions for other toxic elements, including aluminium 88,89 and boron 90 .
Higher atmospheric levels of CO2 and other greenhouse gases are predicted to increase the frequency and duration of heat-waves 91 , which will lead to losses in crop yield—especially in arid regions 92 . Sensitivity to extreme temperatures varies during the plant lifecycle and across species. Low temperatures influence the germination, establishment, growth and viability of crops, except for those with temporal chilling or freezing resilience (such as winter wheat). Genetic variation in key transcriptional regulators of cold resilience is leveraged in breeding of several grain crops 46 . By contrast, warm temperatures promote growth until a threshold is reached above which yields precipitously diminish, especially when soil moisture is low or humidity is high 93,94 . Sensitivity to temperature extremes is heightened during reproduction, when it reduces male fertility and seed quality 95 . This presents a daunting challenge as protective responses are typically accompanied by reduced yield. Heat stress is an expanding threat in tropical regions, because, at high humidity, plants are less able to cool their leaves by transpiration via stomatal pores that control the trade-off between CO2 intake and water loss 96 . There is an urgent need for research and for ensuing genetic and engineered solutions that preserve crop productivity at increased temperatures (Box 1).
Breeding or engineering plants for a high yield potential in varied and variable environments is a potential solution for capturing effective resilience. Plants typically dampen growth and accelerate reproductive development as a consequence of stress. Yield maintenance under moderate drought was significantly improved in AQUAmax corn hybrids produced by selective and marker-assisted breeding 97 . The underlying genetic variants and mechanisms that enable these lines to conserve soil moisture and delay the accumulation of biomass until grain filling remain to be characterized. Higher yields under well-watered conditions as well as under moderate drought at the time of flowering was achieved in corn that expresses a metabolic enzyme that converts the low-abundance metabolite trehalose-6-phosphate (T6P) to trehalose in the phloem companion cells at the base of the ear and developing florets 98 (Fig. 2e). This cell-specific modulation of T6P augments the mobilization of photosynthate to the unfertilized floret, and prolongs the photosynthetic activity of leaves during grain filling. The spatial modulation of T6P levels also regulates the draw of seed reserves into young elongating shoots of rice, particularly when dry seeds are sown directly into a flooded paddy 99 . The application of plant-permeable T6P analogues to wheat leaves increased seed filling and improved recovery from drought 100 . These examples illustrate the critical integration of metabolism and stress resilience to improve crops that can be provided by genetic variation and engineering.
Modern crops are highly efficient at rapidly spreading their leaf canopies to maximize light interception, and at partitioning carbon and nutrients into seeds. However, crops are not as efficient at converting absorbed light energy into sugars through the process of photosynthesis 101 . This may be because the proteins and enzymes that mediate photosynthesis evolved in a low-light marine environment, which was very different from modern agronomic and atmospheric conditions 102 . However, the conservation of chloroplast transmembrane proteins that collect light energy and participate in electron transfer reactions within the chloroplast, along with the conservation of enzymes involved in carbon fixation, reduction and regeneration across plant species 103 (Fig. 2f), has aided the modelling of photosynthesis 104 and identification of numerous targets for increasing its efficiency 101 (Fig. 3). Theoretical targets include expanding and optimizing light capture by the leaf canopy 105,106,107 , inducing a more rapid relaxation of non-photochemical quenching at photosystem II 108 , increasing the carboxylation capacity of the Rubisco enzyme as well as minimizing oxygenation and photorespiration 109 , enhancing the regenerative capacity of the carbon reduction cycle 101 , optimizing the electron transport chain 110 , converting crops from C3 to C4 metabolism 111 , and adding components of cyanobacterial or algal systems to pump CO2 or compartmentalize Rubisco 101 . Improving photosynthetic efficiency is neither a new nor a universally accepted idea. Some have argued that the selection pressures endured by photosynthesis render it unamenable to improvement 112 . Despite decades of research, the challenge of engineering Rubisco for improved specificity and carboxylation rate remains unmet 113 . However, some recent successes in engineering photosynthetic enzymes and introducing novel pathways into chloroplasts may lead to substantial gains in crop performance, as outlined below.
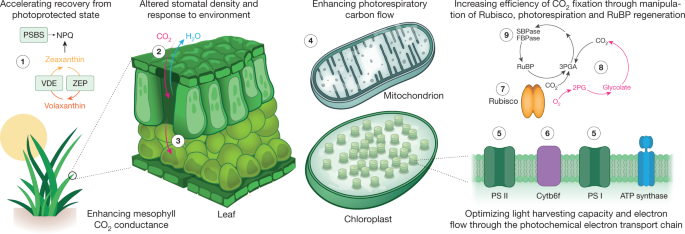
Maize photosynthesis and fresh weight were enhanced by overexpressing the small and large subunits of Rubisco, together with an assembly chaperone protein 114 . In wheat, the overexpression of sedoheptulose-1,7-biphosphatase showed increased photosynthesis, and resulted in increased plant and grain biomass 115 . These genetic modifications to key crops are promising; their ultimate potential can be tested by incorporating the changes into elite varieties, and evaluation in the field. Photosynthetic manipulations also show promise in the field in the model plant tobacco. Re-engineering the expression of enzymes that control the induction, relaxation and amplitude of non-photochemical quenching successfully enhanced photosynthesis during natural light transitions, which resulted in 14–20% greater vegetative biomass in the field 116 . Even greater gains were observed by inserting enzymes involved in glycolate metabolism into chloroplasts to reduce photorespiration 117 . Coupling this with reduced expression of a glycolate and glycerate transporter, to minimize glycolate flux out of the chloroplast, raised vegetative biomass by 40% under field conditions 117 . These studies represent fundamental breakthroughs in understanding and engineering photosynthesis, which can now move from the proof-of-concept stage to expanded field testing.
Crops lose between 100 and >400 water molecules through stomatal pores in leaves for every carbon atom that is fixed by photosynthesis, highlighting a fundamental trade-off between carbohydrate production and water use. Increases in CO2 concentrations inside leaves cause a reduction in the size of stomatal-pore apertures 118 . The continuing rise in atmospheric CO2 is increasingly narrowing stomatal pores, which can enhance the efficiency of water use by crops. However, many crops have weak or non-optimal stomatal CO2 responses. Advances have been made in understanding the signal transduction pathways that regulate water loss in response to CO2 118 , including that the stomatal CO2 response requires amplification by—but also includes unique components upstream of and parallel to—the ABA response pathway in guard cells 119 (Fig. 2g). The upregulation of the stomatal CO2 response by guard-cell-targeted overexpression of carbonic anhydrases increased instantaneous water-use efficiency by about 44% in Arabidopsis, without a reduction in photosynthetic assimilation rates at ambient CO2 120 . On the other hand, C3 crops growing in nutrient-rich and water-sufficient humid regions could benefit from a weaker CO2-induced stomatal closing response, which may enhance growth owing to CO2 ‘fertilization’ in an atmosphere with an increased concentration ofCO2 121 . A complete understanding of the CO2 response pathway is needed to optimize and test water-use efficiency and gas-exchange strategies in the field.
Successful transgenic modifications in barley and rice have shown that reducing the density of stomata improves plant performance under water-restricted conditions 122,123 . Overexpression of the chloroplast photosystem II subunit S protein in tobacco was reported to lower stomatal conductance, and increased the efficiency of water use by field-grown plants 124 . The effective manipulation of stomatal function will require the discovery of the primary CO2 and/or bicarbonate sensors that control the stomatal CO2 response, as well as harnessing natural genetic variation in stomatal properties that could improve trade-offs between carbon gain and water loss in a world with high levels of atmospheric CO2 125 .
Yields of crops are heavily dependent on sufficient nutrition (in particular, nitrogen and phosphorus) that is currently provided primarily through the application of inorganic fertilizers. In smallholder farming systems, crop productivity is limited by the availability of these nutrients 15 . Substantial advances have been made in understanding the mechanisms of nutrient uptake, transport and use in plants, with the aim of improving sufficiency 126 (Fig. 2h). Balancing the use of photo-assimilate with nutrient uptake is critical for optimizing yields. The mutations that confer stem shortening in cereals, which facilitated the Green Revolution, brought with them unintended inefficiencies in nitrogen use that can be compensated for by changing the balance of transcription factors that control growth and nutrient use 127 . Breeding can also contribute to reducing nutrient imbalances through the optimization of rooting systems, nutrient transport activity and partitioning 128 .
In natural ecosystems, plants frequently engage with beneficial microorganisms that facilitate the uptake of limiting nutrients such as nitrogen and phosphate 129 . In agriculture, these beneficial associations are often dampened by supplied fertilizers, because plants suppress their interaction with symbionts when they perceive ample nutrients. Most plant species associate with arbuscular mycorrhizal fungi that greatly expand the root-surface area for nutrient uptake, and which actively mine immobilized phosphates from the soil 130,131 . Bringing associations with arbuscular mycorrhizal fungi more effectively into annual cropping systems with moderate fertilizer use could improve nutrient capture, and increase sustainability—particularly if the phosphate suppression of mycorrhization could be overridden. However, applications of strigolactones (the plant-derived low-phosphate signals to microorganisms) have so far been insufficient to override the suppression of mycorrhization 132 and more research is therefore needed to obtain benefits from mycorrhizal associations in agriculture.
Some plants are colonized intracellularly by nitrogen-fixing bacteria that can deliver the complete nitrogen needs of the host plant. Associations such as this are limited to a select group of species, which presents an opportunity to radically improve nitrogen availability for cereal crops if the symbiosis trait can be transferred. Multiple avenues are being explored to achieve this—from equipping plants to associate with nitrogen-fixing bacteria to the transfer of the enzyme nitrogenase, which is responsible for nitrogen fixation. Studies of these processes in their native context have provided an understanding that was absent 30 years ago, when such approaches were first broached. Evolutionary, genomic and mechanistic studies suggest that relatively few genetic components might be needed to confer nitrogen-fixation capabilities. In the case of transferring nitrogenase to plants, the restriction of genetic components required was achieved by concatermerizing bacterial genetic units to create a minimal set of three genes that are necessary for the transfer of nitrogen fixation 133 . Moreover, some components of nitrogenase can be stably expressed in yeast and plants 134 .
The evolution of the nitrogen-fixing symbiosis in legumes used many components that function in associations with arbuscular mycorrhizal fungi 129 , which means that cereals possess some of the necessary building blocks and have the potential to streamline engineering efforts to transfer the nitrogen-fixing symbiosis. Recent phylogenomic approaches indicate that very minimal gene reduction (between two and seven genes) is associated with the loss of nitrogen fixation 135,136 , suggesting that a small set of genes could convert a species that associates with arbuscular mycorrhizal fungi into one that can also form nitrogen-fixing symbiosis. This considerable engineering challenge will require precise transcriptional and post-translational regulation of multiple heterologous genes in cereals. Additional challenges are the few well-characterized promoters for gene regulation in cereal roots, and bottlenecks associated with transformation of cereals that limit the scale of throughput required to test the engineering iterations that will be necessary to achieve nitrogen fixation.
The environment around and within plant roots includes microbial communities. These can be relatively restricted 137 or dynamic 138 , and responsive to nutrient status 139 . Such communities or community members have the potential to protect plants against pathogen infections 140,141 and, to some extent, drought 142,143 . A greater understanding of the mechanisms and the environmental conditions, including climate effects 144 , that control plant–microorganism assembly and activities may enable the engineering of microbial communities to optimize crop performance, particularly with microorganisms that are engineered using synthetic biology approaches. Current research indicates that some fungal species benefit host plants by enhancing phosphate uptake 145 , and within the diversity of cereal crops are lines that can host active communities of nitrogen-fixers 146 . The manipulation of microbial associations to improve crop resilience to environmental stresses is an area of intense research.
Research advances have provided innovative opportunities and technologies across the plant sciences, which can furnish solutions for addressing future food security (Box 1). The strategies described here for enhancing the resilience and sustainability of crops will only be realistic if they are part of an integrated approach to agriculture that is developed collaboratively with agronomists, engineers and farmers 147 . A critical challenge is the time from research discovery to true and widespread implementation in agriculture. Some high-impact breeding and genetically modified traits (for example, pest resistance mediated by individual Bacillus thuringiensis Cry proteins) have spread relatively rapidly. However, even in cases that involve breeding into diverse varieties, the time from initial discovery and development to broad use has often exceeded ten years 79 . Regulatory processes and intellectual property hurdles associated with technology can lead to additional delays in implementation. The robust assessment of varieties in variable field environments is essential to timely adoption. In the case of submergence-tolerant SUB1 rice varieties 50 , cooperation between scientists, breeders and farm advisers helped to achieve farmer acceptance and governmental certification within three years of gene characterization. The visible yield advantage of SUB1 varieties after flooding, and their lack of differences with the varieties they replaced, was key to their adoption. This success contrasts with the failure to provide many farmers in climate-vulnerable areas with the services of plant breeders to mobilize genetic variation for crop improvement 148 and with selected or engineered genotypes that did not translate to the field 46 . Complementary approaches and technologies may provide viable opportunities (such as high-protein, salt-tolerant algae that require limited freshwater)—although new infrastructure, energy inputs and engineering solutions will be needed 149 .
Valuable genetic diversity for increasing crop resilience resides in cultivated landraces, heirloom varieties and the wild relatives of crops. Seed banks curate and distribute crop germplasm; the Crop Trust (https://www.croptrust.org) is one of the leading efforts to collect, conserve and use the approximately 50,000 species of wild relatives of crops 150 . These seed banks distribute germplasm that can be tapped for adaptations to abiotic and biotic stresses, but greater investment in high-throughput genotyping and phenotyping is needed to accelerate mapping, the identification of genes and mechanisms, and downstream breeding 151 .
Addressing yield loss due to climate change, salinity and (re)emerging diseases, weeds, parasitic plants and pests requires innovative technologies and proactive responses, not unlike the development of vaccines and innovations in modern medicine. The integration of genetic resources and transformative technologies, from genome editing to synthetic biology, are necessary to capture traits that increase global food security and reduce the effects of agriculture on the environment. An early failure of plant biotechnologists was in the lack of effective engagement with environmentalists, farmers and consumers on questions of health and safety, despite strict governmental procedures for the validation, release and monitoring of genetically modified crops. It is critical that the specific method used for crop improvement does not stymie the implementation of safe and effective solutions. Non-politicized regulatory systems are essential for scientific advances to scale to farmers within the timeframe needed.
The current timeline for increasing the resilience and sustainability of crops is too long. Crop varieties with new combinations or variants of disease-resistance genes are in preparation for use against newly emerged virulent pathogens. Advances in sequencing and the early detection of invasive pathogenic strains should enable better monitoring of disease, and therefore knowledge regarding where to deploy particular crop genotypes. The horizon for tailored panels of appropriately controlled genes that impart functional immunity in a commercial crop is years away. The most rapid translation to the field will be for small suites of genes from existing crop germplasm. For challenges that are difficult to overcome (such as resilience to heat and aridity during plant sexual reproduction 98 ), disruptive advances such as the asexual propagation of seeds 152 could lessen yield loss due to male infertility. Success in the engineering of improved photosynthesis, nutrient use and beneficial plant–microorganism interactions requires intensive investment, but could result in the gains needed.
The plant sciences have a critical role in meeting the food and fibre challenges of the future. Timely investments and research at many levels and collaborative efforts are paramount to deploying resilience mechanisms and improving the sustainability, yields and nutritional value of our crops.
We apologize to those authors whose research could not be cited owing to space limits. We thank P. J. Franks and E. Buckler for discussions, and A. Digrado and C. Benjamin for assistance with figures. Research in the authors’ laboratories was supported by grants from the US National Science Foundation to J.I.S. (MCB-1900567) and J.B.-S. (IOS-1546879; IOS-1810468; IOS-1856749); the US National Institutes of Health to J.I.S. (GM060396-ES010337); the National Institute of Food and Agriculture to J.B.-S. (2017-67013-26194; 2019-67013-29313); the Max-Planck Society and the Deutsche Forschungsgemeinschaft (DFG; German Research Foundation) under Germany’s Excellence Strategy (EXC-2048/1, project 390686111 and Priority Program 2125 ‘DECRyPT’) to J.E.P.; a sub-award to E.A.A. from the University of Illinois as part of the Realizing Increased Photosynthetic Efficiency (RIPE) project (OPP1060461), UK AID and the Foundation for Food and Agricultural Research; and the Engineering the Nitrogen Symbiosis in Africa project (OPP1172165) sponsored by the Bill & Melinda Gates Foundation and Gatsby Foundation grants to G.E.D.O.